Defines a new alias, region, grid, axis, variable, or viewport.
DEFINE ALIAS
Defines an alias for a command. "ALIAS" is an alias for DEFINE ALIAS.
yes? DEFINE ALIAS NAME COMMAND
Example:
yes? DEFINE ALIAS SDF SHOW DATA/FULL
DEFINE ATTRIBUTE
/DATASET /TYPE /OUTPUT
Defines a new attribute for a variable. The variable may be from a netCDF dataset, a user-defined variable, or a variable from another type of dataset. (See the section on access to dataset and variable attributes.)
yes? DEFINE ATTRIBUTE[/qualifiers] varname.attname = expression
Example: add new attributes with DEFINE ATTRIBUTE
yes? USE etopo60
yes? DEFINE ATT rose.floatval = 22
yes? DEFINE ATT rose.pp = {1.5, 1.9}
yes? DEFINE ATT/TYPE=string/OUTPUT rose.strval = "some text about ROSE"
yes? DEFINE ATT/TYPE=string/OUTPUT rose.str_att = 1234.5
yes? DEFINE ATT/TYPE=int/OUTPUT rose.ipp = {1, 273, 20, 70}/DATASET=n applies the attribute to the variable in the specified dataset.
/TYPE is any of the the NetCDF types STRING, DOUBLE, FLOAT, INT, SHORT, and BYTE. If it is not specified, it is inferred from the expression. (Previous to Ferret v7, only FLOAT and STRING were allowed). If /TYPE=string is given with a numeric value to the right of the equals sign, the output will be converted to to a string type attribute. Lists of strings are not allowed. The default type for numeric attributes is float.
/OUTPUT sets the flag for output so that this attribute will be written when the variable is written to a netCDF file. By default, when an attribute is defined this flag is marked for output. Use CANCEL ATTRIBUTE/OUTPUT to mark an attribute as not-written.
NOTE: A restriction on defining attributes, is that the special attributes scale_factor and add_offset may be defined in Ferret only for user-defined variables not variables from files. These attributes affect the reading of data from netCDF files and so are special. If these attributes already exist in a netCDF file, they will automatically be written, if writing, say a subset to a new file. To add these attributes to a variable for writing or to change the values of these attributes, define a new variable
yes? use coads_climatology yes? set var/name=sst_in sst yes? let/title="SST"/units="Deg C" sst = sst_in yes? define att/output sst.scale_factor = 0.01 yes? define att/output sst.add_offset = 10 yes? set var/bad=-3000/outtype=short sst yes? save/clobber/file=sst.nc/x=0/l=1 sst
DEFINE AXIS
/X/Y/Z/T/E/F /DEPTH /FILE /FROM_DATA /LIKE /MODULO /NAME /NPOINTS /T0 /UNITS /EDGES /CALENDAR /BOUNDS
Defines an axis (axis name up to 16 characters). The syntax is:
yes? DEFINE AXIS[/qualifiers] axis_name or yes? DEFINE AXIS[/qualifiers] axis_name = expr
Example:
yes? DEFINE AXIS/X=140E:140W:.2 AX140
or
yes? DEFINE AXIS/X myaxis = {1, 5, 15, 35}
Axes which are "in use", because they are used by currently open data sets may be redefined using DEFINE AXIS. (In early versions of Ferret, attempting to redefine an in-use axis generated an error.)
This feature is especially useful to correct the interpretation of erroneous files, or files which exhibit minor incompatibilities with Ferret. Use this feature with caution as it can be used to "fool" Ferret into an incorrect interpretation of a data file.
Command qualifiers for DEFINE AXIS:
Specifies the limits and point spacing of an axis.
yes? DEFINE AXIS/X=lo:hi:delta axis_name
The limits may be in longitude, latitude, or date format (for X, Y, or T or F axis, respectively) or may be simple numbers. No units are assumed unless units are given explicitly with the /UNITS qualifier.
Use /UNITS=degrees to obtain latitude or longitude axes. The X or Y qualifier determines which orientation "degrees" refers to. If an X axis has units of degrees longitude it is automatically marked as a modulo axis.
For T or F axes, the limits may be dates (dd-mmm-yyyy:hh:mm:ss) or may be time steps. The delta increment is regarded as hours unless the /UNITS qualifier specifies otherwise. Note that time axes may not extend beyond year 9999. If an F axis has units of time, it is treated as the Forecast axis for a Forecast Model Run Collection. (See documentation by searching for FMRC at the Unidata site, for instance.)
If the time limits are given as dates then this axis produces date-formatted output (unless CANCEL MODE CALENDAR is issued). If the time limits are given as time steps then all instances of this axis are labeled with time step values in the units specified with the /UNITS qualifier.
Examples (evenly-spaced axes):
yes? DEFINE AXIS/X=140E:140W:.2 ax140 yes? DEFINE AXIS/Y=15S:25N:.5 axynew yes? DEFINE AXIS/Z=0:5000:20/UNITS=CM/DEPTH axzcm yes? DEFINE AXIS/T="7-NOV-1953":"23-AUG-1988:11:00":24 axtlife yes? DEFINE AXIS/T=0:30:1/t0=1-jan-2010/units=day tday yes? DEFINE AXIS/T=25:125:5/UNITS=minutes axt5min
Allows for non-Gregorian calendar axes. The calendars allowed are
|
calendar name |
number of days/year |
notes |
|
GREGORIAN or STANDARD |
365.2425 |
default calendar: proleptic Gregorian |
|
JULIAN |
365.25 |
with leap years |
|
NOLEAP |
365 |
no leap years |
|
ALL_LEAP |
366 |
366 days every year |
|
360_DAY |
360 |
each month is 30 days |
The calendar definitions conform to the netCDF conventions document for calendars. See the section about calendars in Chapter 4 of the CF document at http://cfconventions.org/. These calendar definitions are compatible with the Udunits standard (see http://www.unidata.ucar.edu/software/udunits/udunits-1/udunits.txt ) which has slightly different naming conventions. Also note that the default calendar in Ferret is the proleptic Gregorian calendar, i.e. the definition of a year is consistent throughout time and does not have an offset in the 1500's as the historical calendars did. However, files written using the NOAA/CDC standard for the "blended" Julian/Gregorian calendar are read correctly by Ferret: If a time axis has a time origin of 1-1-1 00:00:00, and uses the default calendar, and if the coordinates of axis lie entirely after the year 1582, then the historical 2-day shift is applied.
The netCDF conventions recommend that the calendar be specified by the attribute time:calendar when there is a non-Gregorian calendar associated with a data set, i.e.
time:calendar=noleap
Ferret reads this attribute from a netCDF file, or gets it from a definition made with DEFINE ATTRIBUTE and assigns the designated calendar to the time axis.
Example:
Define a calendar axis and regrid an existing variable to this axis. (Use the continuation character \ to break this long line into two lines.)
yes? DEFINE AXIS/CALENDAR=JULIAN/T="15-JAN-1982":"15-DEC-1985":30\ /UNITS=days tmodel yes? LET twind = uwnd[GT=tmodel@NRST]
When regridding from one calendar axis to another the length of a year is assumed to be constant, therefore the regridding calculates a scale factor based on the length of a second in each calendar, computed from the number of seconds per year for the calendars.
Specifies the Z axis to be a depth, positive downward, axis. A depth axis is indicated by a "(-)" following its title in a SHOW GRID or SHOW AXIS command. Depth axes are notated by "UD" (up-down) in the grid definition file, while normal vertical axes (such as an elevation axis in meteorology) are notated by "DU" (down-up).
Example:
yes? DEFINE AXIS/Z=0:5000:20/DEPTH/UNITS=CM AXZDCM
The /EDGES qualifier indicates that the coordinates provided refer to the edges or boundaries between grid cells. When /EDGES is used, the coordinates of the grid points will be computed at the midpoints between the indicated edges. When /EDGES is used in conjunction with /FROM_DATA the number of grid points created will be equal to the number of coordinates minus one, since the list of edges includes both the upper and lower edge of the axis. An example of defining an axis by its edges is
yes? DEFINE AXIS/Z=0:5010:20/EDGES/DEPTH/UNITS=CM AXZDCM
A class of especially important uses for the /EDGES qualifier is to create custom calendar axes. This example creates a true monthly axis, with axis cells beginning on the first of each month (on a Gregorian calendar; see the note below):
yes? ! Define a 20 year monthly axis starting in Jan 1950 yes? LET start_year = 1950 yes? LET nyears = 20 yes? LET indices = L[L=1:`nyears*12`] yes? LET month = MOD(indices-1,12)+1 yes? LET year = start_year + INT((indices-1)/12) yes? DEFINE AXIS/UNITS=days/T0=1-jan-1900/EDGES truemonth = DAYS1900(year,month,1) yes? ! Examine one year of it yes? SHOW AXIS/T=1-jan-1958:1-jan-1959 truemonth name axis # pts start end TRUEMONTH TIME 239 i 16-JAN-1950 12:00 16-NOV-1969 00:00 T0 = 1-JAN-1900 Axis span (to cell edges) = 7274 L T TBOX TBOXLO TSTEP (DAYS) 96> 16-DEC-1957 12:00:00 31 01-DEC-1957 00:00:00 21168.5 97> 16-JAN-1958 12:00:00 31 01-JAN-1958 00:00:00 21199.5 98> 15-FEB-1958 00:00:00 28 01-FEB-1958 00:00:00 21229 99> 16-MAR-1958 12:00:00 31 01-MAR-1958 00:00:00 21258.5 100> 16-APR-1958 00:00:00 30 01-APR-1958 00:00:00 21289 101> 16-MAY-1958 12:00:00 31 01-MAY-1958 00:00:00 21319.5 102> 16-JUN-1958 00:00:00 30 01-JUN-1958 00:00:00 21350 103> 16-JUL-1958 12:00:00 31 01-JUL-1958 00:00:00 21380.5 104> 16-AUG-1958 12:00:00 31 01-AUG-1958 00:00:00 21411.5 105> 16-SEP-1958 00:00:00 30 01-SEP-1958 00:00:00 21442 106> 16-OCT-1958 12:00:00 31 01-OCT-1958 00:00:00 21472.5 107> 16-NOV-1958 00:00:00 30 01-NOV-1958 00:00:00 21503 108> 16-DEC-1958 12:00:00 31 01-DEC-1958 00:00:00 21533.5 109> 16-JAN-1959 12:00:00 31 01-JAN-1959 00:00:00 21564.5
The example above uses the DAYS1900 function, which returns days according to the default Proleptic Gregorian calendar. For other calendars, there is a script def_monthaxis_days.jnl, a contribution from a user.
yes? GO/HELP def_monthaxis_days yes? GO def_monthaxis_days.jnl NOLEAP 2002 2005 mytaxis
These could be modified to define, for instance, a seasonal average, by changing the definitions as follows:
yes? ! Define a 20 year monthly axis starting in Jan 1950 yes? LET start_year = 1950 yes? LET nyears = 20 yes? LET indices = L[L=1:`nyears*4`] ! Starting month of every three month period 1,4,7,10... (centered on 2,5,8,11 Seasons JFM, etc) yes? LET month = MOD(INDICES*3-3,12)+1 yes? LET year = start_year + INT((indices-1)/4) yes? DEFINE AXIS/UNITS=days/T0=1-jan-1900/EDGES three_month_axis = DAYS1900(year,month,1) ! Now try it with LET month = MOD(INDICES*3-3,12)+2 for 3-month intervals FMA, etc
The following example shows the computation of a custom climatological average. Given, for example, a multi-year time series of a daily measured variable, the climatological average of the variable for two unequal time periods could be computed by creating an axis with two points, using the FROM_DATA qualifier. The grid cells for these two points would extend from 15-Mar to 27-May (about 73 days), and from 27-May to 15-Mar (about 292 days). The actual dates on which the 2 points are located would be the midpoints of these two intervals, on 20-Apr and 20-Oct.
yes? DEFINE AXIS/t=1-jan-0001:1-jan-0002:1/unit=days/t0=1-jan-0000 tencoding
yes? LET tstep = t[gt=tencoding]
yes? LET start_date = tstep[t=15-mar-0001]
yes? LET end_date = tstep[t=27-may-0001]
yes? DEFINE AXIS/T/UNITS=days/T0=1-jan-0000/EDGES/MODULO \
tax={`start_date,p=7`,`end_date,p=7`,`start_date+365.2425,p=7`}
yes? DEFINE GRID/T=tax taxgrid
yes? SHOW/L=1:2 grid taxgrid
GRID TAXGRID
name axis # pts start end
normal X
normal Y
normal Z
TAX TIME 2mi 20-APR 12:00 20-OCT 02:54
L T BOX_SIZE TIME_STEP (DAYS)
1> 20-APR 12:00:00 73 475.5
2> 20-OCT 02:54:35 292.2425 658.1212
Define an axis from lists of coordinates and bounds. (See the discussion of netCDF bounds.) The specificaion may be either in terms of 2*N bounds or N+1 edges. For 2*N bounds the upper bound of each cell must be the same as the lower bound of the next cell.The coordinates must be inside (or may coincide with) the cell bounds. (Spaces are used here to clarify the pairs of bounds, but are not necessary and do not affect the way the data is read.)
DEFINE AXIS/X/BOUNDS axisname = coordinates, bounds
Examples:
First, a 2*N bounds definition and then an N+1 one. Note that the second definition has the coordinate z=0 at the same location as the edge of the first grid cell.
yes? DEF AXIS/X/BOUNDS xax = {1,2,5,6}, {0.5,1.5, 1.5,2.5, 2.5,5.5, 5.5,6.5}
yes? LET a = {0,20,50,75,120}
yes? LET b = {0,10,30,60,90,150}
yes? DEF AXIS/Z/DEPTH/BOUNDS zax = a, b
Reads a gridfile for grid and axis definitions. The gridfile specified should be in the standard TMAP gridfile format. There are several documents in $FER_DIR/doc regarding gridfiles and TMAP format (e.g., "about_grid_files.txt").
yes? DEFINE AXIS/FILE=grid_file.grd
Used only in conjunction with /NAME to define an axis from any expression that Ferret can evaluate.
yes? DEFINE AXIS/FROM_DATA/NAME=axis_name expr
(This is a mechanism to convert dependent variables into independent axis data.)
When defining an axis from a LET-defined variable or expression the condensed syntax (e.g.)
yes? DEFINE AXIS/X axname=expression
replaces the older (still supported) syntax
yes? DEFINE AXIS/X/NAME=axname/FROM_DATA expression
Note that the values from which the axis is to be created must be in strictly increasing order. If the coordinates are repeated, Ferret will "micro-adjust" the values by adding multiples of 1 millionth of the axis range to the repeated values. Ferret will issue an informative message if it is micro-adjusting an axis.
Example (unevenly-spaced axes):
yes? DEFINE AXIS/X my_xaxis=pos[D=2]^0.5
yes? DEFINE AXIS/Z/DEPTH/UNITS=meters zdepth={0,10,20,50,100,270}
The first defines each coordinate of the axis "my_xaxis" as the square root of variable "pos" from data set 2. The second defines the axis from a list of numbers. See also DEFINE AXIS/BOUNDS for syntax to define the cell edges as well as coordinate values.
In Ferret v6.8 and higher, variables and coordinate data are both represented within Ferret as double precision numbers. The next section applies only to versions of Ferret previous to Ferret v6.8:
Variables in Ferret are always represented as single precision floating-point numbers. Coordinates are represented as double precision values. This presents a quandary when we wish to define an axis, and have double precision data representing the coordinates available as a variable (but not a coordinate variable) in a netCDF file. Once that variable is read by Ferret, it becomes single precision and we lose the precision needed to define the axis. Starting with V6 of Ferret, a means is provided to work around this, in particular for making graphics. We read the data, specifying an offset which is applied when the data is read, and which gives the numbers fewer significant figures, so the variations are represented in single precision. This may be "undone" via a PPLUS command which applies the reverse offset to the coordinates at the time we make a plot.
For example, say a variable called XVALS is in a netCDF file, which has data values {144.002, 144.0034, 144.005}. We want to create an axis from these values, and put the variable data_var onto that axis. Once XVALS is converted to single precision we would lose the variation. We can read the locations with the SET VAR/OFFSET command. It is convenient to define a variable to keep the constant offset, as it will be used again.
Example:
yes? SET DATA filename.nc yes? LET xlon = 144 yes? SET VAR/OFFSET=`-1*xlon` xvals yes? ! when the data is read, the offset is added yes? LIST/NOHEADER xvals 1 / 1: 0.002000 2 / 2: 0.003400 3 / 3: 0.005000 yes? ! Define an axis, and put the variable onto this axis... yes? DEFINE AXIS/X/UNITS=deg xxaxis = xvals yes? LET var = data_var[gx=xxaxis@ASN] yes? ! Add back the offset. This works with any plotting command. yes? PLOT/SET var yes? PPL XVALOFF `xlon` yes? PPL PLOT
(Ferret v7.4) Specifies that the new axis will have the same attributes as an existing defined axis.
The DEFINE AXIS/LIKE= command must include a direction, /X= /Y= etc. The direction must match the direction of the axis whose attributes we are inheriting. The coordinates are not inherited; only the attributes, such as units. For time axes the units and time-origin are inherited, as well as the calendar definition (e.g. non-Gregorian calendars such as noleap). For vertical axes, the positive=up or down attribute is inherited.
If qualifiers such as /UNITS or /CALENDAR are given on the command line those settings supersede the inherited attribute.
Specifies that the axis being defined be treated as modulo; that is, the first point will wrap around and follow the last point (e.g., a longitude axis).The optional modulo length is the length in axis units of the modulo repeat. If a length is specified, it may be longer than the axis span, so that the axis is treated as a subspan modulo axis, and if no length is specified then the default modulo length for the type of axis is used. See the sections on modulo axes and subspan modulo axes for more information
Used only in conjunction with /FROM_DATA to specify the name of the axis to be defined.
yes? DEFINE AXIS/FROM_DATA/NAME=axis_name expr
Specifies the number of coordinate points on the axis being defined.
yes? DEFINE AXIS/Z=lo:hi/NPOINTS=n ax_name
This qualifier can be used instead of specifying Z=lo:hi:delta.
Specifies the date and time associated with the time step value 0.0, for a T or an F axis.
Example:
DEFINE AXIS/T="1-NOV-1980":"15-AUG-1988":72/T0="1-JAN-1800" TNEW DEFINE AXIS/F="1-JAN-2001":"15-JAN-2001":1/T0="1-JAN-2010"/UNITS=days FORECAST_TIME
Note: The /T0 qualifier is optional; the underlying time step values are transparent to Ferret users for most purposes. The default value is 15-JAN-1901.
Specifies the units of the axis being defined.
A DEFINE AXIS command such as DEFINE AXIS/X=130E:80W:2 xax infers from the formatting of the longitude coordinates the implied qualifier "/UNITS=degrees". Similar for latitudes.
Example:
yes? DEFINE AXIS/Z=0:2000:100/UNITS=CM ZCM
Any string (up to 10 characters) is acceptable as a units string, but only the following units are recognized and used when computing axis transformations:
|
cm (or centimeter) |
mm (or millimeter) |
day |
|
km (or kilometer) |
mb (or millibar) |
mon |
|
m (or meter, or metre) |
level |
yr (or year) (default Proleptic Gregorian year) |
|
deg (or lat or lon) |
layer |
|
|
ft (or feet or foot) |
sec |
year360 (360 days) |
|
in |
min |
year366 (366 days) |
|
mile |
hour |
M2 cycles |
|
dbar |
mbar |
NOTES:
1) As of Ferret version 5.1 the definition of the unit "month" has been redefined to be exactly 1/12 of a climatological year. This change applies both to files that use "units=months" and to the DEFINE VARIABLE command. The climatological month is the length of an average month in the Gregorian calendar, including leap years -- 1/12 or 365.2485 days. Thus the command
yes? DEFINE AXIS/T0=1-JAN-0000/T=0:12:1/EDGES/units=months/MODULO month_reg
defines a climatological monthly axis which does not "drift" over time due to leap years. This non-drift behavior can be observed using a commands like
yes? SHOW AXIS /l=1:12001:1200 month_reg
which will show every 100th January over 1000 years. To create a monthly Gregorian calendar axis - with axis cells beginning on the first of each month, see the example under DEFINE AXIS/EDGES .
2) The units dbar and mbar are recognized by Ferret, however, no automatic conversion is attempted between these and any other units.
Ferret will convert recognized units of length to meters and recognized units of time to seconds during transformations such as integration (@IIN and @DIN) and differentiation (@DDB, @DDC, @DDF) (see "General Information about transformations"). Using this characteristic it is always possible to query Ferret about the conversion factors from meters or seconds by integrating a grid cell of width one on an axis of the units in question. For example:
yes? ! query conversion factor to meters yes? define axis/x=0:1:1/edges/units=feet xtest ! 1 point, cell width=1 unit yes? let vx = 0*X[gx=xtest]+1 ! vx = 1 yes? list/prec=7 vx[x=@din] 0*X[GX=XTEST]+1 X (FEET): 0 to 1 (integrated) 0.3048000 yes? ! query conversion factor to seconds yes? define axis/t=0:1:1/edges/units=month ttest ! 1 point, cell width=1 unit *** NOTE: /UNIT=MONTHS is ambiguous ... using 1/12 of 365 days. yes? let vt = 0*T[gt=ttest]+1 ! vt = 1 yes? list/prec=7 vt[t=@din] 0*T[GT=TTEST]+1 T (MONTH): 0 to 1 (integrated) 2628000.
2) If an F axis has any of the units of time, Ferret will treat it as the Forecast axis for a Forecast Model Run Collection. (See documentation by searching for FMRC at the Unidata site, for instance.)
DEFINE GRID
/X/Y/Z/T/E/F /FILE /LIKE
Defines a grid (name may be up to 16 characters).
yes? DEFINE GRID[/qualifiers] grid_name
Example:
yes? DEFINE GRID/LIKE=temp/T=my_t_axis my_grid
Command qualifiers for DEFINE GRID:
Specifies what particular axis is to be the X, Y, Z, T, E, or F axis for this grid.
yes? DEFINE GRID/X=axname grid_name
The name axname may be the name of an axis, the name of a grid that uses the axis desired, or the name of a variable for which the defining grid uses the axis desired.
For example,
yes? DEFINE GRID/X=U gx
will create a grid named gx which is one-dimensional—normal to Y, Z, T, E, and F.
Note: Many axes possess an orientation implicit in their units, especially latitude, longitude, and time axes. The effects of using an axis in an inappropriate orientation, such as /X=time_axis, are unpredictable.
[obsolete: This capability is no longer maintained.] Reads a gridfile for GRID and AXIS definitions. The gridfile specified should be in the standard TMAP gridfile format. There are several documents in $FER_DIR/doc regarding gridfiles and TMAP format (e.g., about_grid_files.txt).
Example:
yes? DEFINE GRID/FILE=new_grids.grd
Specifies a particular grid (by name or by reference to a variable defined on that grid) to use as a template to create a new grid.
yes? DEFINE GRID/LIKE=grid_or_variable_name grid_name
All axes of the grid being created will be identical to the axes of the "LIKE=" grid except those explicitly changed with the /X, /Y, /Z, /T, E, or F qualifiers. The argument may be an expression.
Example:
yes? DEFINE GRID/LIKE=temp[D=2]/Z=ZAX gnew !temp from data set 2
Examples: DEFINE GRID
1) yes? DEFINE AXIS/T="1-JAN-1980":"31-DEC-1983":24 axday
yes? DEFINE GRID/LIKE=temp/T=axday gday
Define grid gday to be like the defining grid for temp but with a 4-year, daily-interval time axis.
2) yes? DEFINE GRID/LIKE=temp[D=ba022]/T=sst[D=nmc] gnmc3d
Define grid gnmc3d like temp from data set ba022 but with the same time axis as sst from data set nmc.
3) yes? DEFINE AXIS/X=140E:140W:.2 xnew
yes? DEFINE AXIS/Y=5S:5N:.2 ynew
yes? DEFINE AXIS/T="15-FEB-1982":"15-FEB-1984":48 tnew
yes? DEFINE GRID/X=xnew/Y=ynew/T=tnew gnew
Define grid gnew from new axes. The grid, gnew, will be normal (perpendicular) to Z.
DEFINE REGION
/I/J/K/L/M/N /X/Y/Z/T/E/F /DI/DJ/DK/DL/DM/DN /DX/DY/DZ/DT/DE/DF /DEFAULT
Defines or redefines a named region_name (first 4 characters are recognized).
yes? DEFINE REGION[/qualifiers] region_name
If the qualifier /DEFAULT is not given only those axes explicitly named will be stored. If the qualifier /DEFAULT is given all axes will be stored.
Command qualifiers for DEFINE REGION:
DEFINE REGION/I=/J=/K=/L=/M=/N=/X=/Y=/Z=/T=/E=/F=
Specifies region limits ( =lo:hi or =val).
DEFINE REGION/DI=/DJ=/DK=/DL=/DM=/DN=/DX=/DY=/DZ=/DT=/DE=/DF=
Specifies a change in region relative to the current settings (=lo:hi or =val). See examples below.
Saves all axes and transformations, not just those explicitly given. Commonly, a GO script begins with "DEFINE REGION/DEFAULT save" and ends with "SET REGION save".
Examples: DEFINE REGION
1) yes? DEFINE REGION/DEFAULT save
Stores the current default region under the name "save". The region may be restored at a later time by the command yes? SET REGION save.
2) yes? DEFINE REGION/X xonly
Stores the current default X axis limits, only, as region xonly.
3) yes? DEFINE REGION/DX=-5 xonly
Stores the current default X axis limits minus 5 as region xonly.
4) yes? DEFINE REGION/Y=20S:20N/Z yanz
Stores the given limits from the Y axis and the default Z axis limits.
5) yes? DEFINE REGION/DEFAULT/L=5 l5
Stores the current default region with the modification that L, the time step, is stored as 5.
6) yes? DEFINE REGION/DL=-1:+1 lp2
Stores an L region beginning 1 time step earlier and ending 1 time step later than the current default region as region lp2.
DEFINE SYMBOL
Allows the user to define a string variable. Symbol names must begin with a letter and contain only letters, digits, underscores, and dollar signs.
yes? DEFINE symbol symbol_name=string
Example:
yes? DEFINE symbol my_x_label = sample number
The total number of symbols is limited to 5000; including those defined by DEFINE SYMBOL commands and those automatically defined by Ferret and PPLUS, which are discussed in the section on special symbols.
DEFINE VARIABLE
/BAD= /D /LIKE= /TITLE= /UNITS=
Allows the user to define a variable from a valid algebraic expression. Note: LET is an alias for DEFINE VARIABLE.
yes? DEFINE VARIABLE[/qualifiers] name=expression
Example:
yes? LET SPEED = U^2 + V^2
Parameters
The expression may be any valid expression. See the chapter "Variables and Expressions", section "Expressions" for a definition of valid expressions.
The name specified with DEFINE VARIABLE can be 1 to 128 characters in length—letters, digits, $ and _, beginning with a letter.
Pseudo-variable names (I, J, K, L, _M, _N, X, Y, Z, T, _E, _F, XBOX, YBOX, ZBOX, TBOX, etc) and operators (AND, OR, GT, GE, LT, LE, EQ, NE) are reserved and cannot be used. See the FAQ about Reserved Keywords for tips on handling datasets which use reserved words as variable names. It is not recommended to use function names such as SIN, EXP, LN or Ferret operators and keywords like PLOT or AXIS as variable names. See the chapter "Variables and Expressions" for recognized operators and functions, or use the commands SHOW COMMAND and SHOW FUNCTION for a list of all commands and functions. The name that you give to the variable is in general not treated in a case-sensitive way. If you define for instance, variables named TEMP and Temp, both in the same Ferret session, the second definition will supersede any previous definitions. The only detail of case-sensitivity that is implemented is that on saving variables to a NetCDF file, the original upper- or lower-case spelling of the variable name will be retained.
If the name defined is the same as a variable name in a data set, the user-defined variable is used instead of the file variable.
Examples:
yes? DEFINE VARIABLE sum = a+b ! or equivalently yes? LET sum = a+b yee? ! Define velocity in m/sec from position, pos, in cm. yes? LET/TITLE="velocity"/UNIT="m/sec" pos[T=@DDC]*0.01
use of the $ character invariables names is legal but is not recommended as any variable names containing $ must be referred to in quotes, e.g.
yes? LET A$B = 7 yes? list 'A$B'
Command qualifiers for DEFINE VARIABLE:
Allows user to control the missing value of user-defined variables The specified value will be used whenever the variable is LISTed (or SAVEd) to a file. Note that the missing value will revert to its default (-1E34) when this variable is combined in further calculations.
Example:
yes? let/bad=3 gap_3 = I[I=1:5] yes? list gap_3 I[I=1:5] 1 / 1: 1.000 2 / 2: 2.000 3 / 3: .... 4 / 4: 4.000 5 / 5: 5.000 yes? let new_var = gap_3 + 5 yes? list new_var GAP_3 + 5 1 / 1: 6.00 2 / 2: 7.00 3 / 3: .... 4 / 4: 9.00 5 / 5: 10.00 yes? list/form=(1PG15.3) new_var GAP_3 + 5 X: 0.5 to 5.5 6.00 7.00 -1.000E+34 9.00 10.0
DEFINE VARIABLE/LIKE=var
(Ferret v7.4) When the variable is defined, it inherits the attributes from an existing variable. This existing variable may be a file variable or a user-defined variable. The new variable will have attributes such as units and title automatically defined from the existing one. The exception is the missing-value attribute. This will not be inherited but will be the default bad-value of -1.e34 or will be set by a /BAD= qualifier.
In addition if the source variable is a file variable, then any or scale_factor or add_offset attributes will NOT be inherited.
Examples:
! Define a surface temperature from an ocean temperature variable. The units
! of "deg C" and the title of "Temperature" are automatically given to sst.
yes? use ocean_temp_data
yes? let/like=temperature sst = temperature[z=0:5@MAX]
yes? list sst[x=300,y=-20]
VARIABLE : Temperature (deg C)
...
! To use a different title, simply specify that on the command line.
yes? let/like=temperature/title="Surface Temperature" sst = temperature[z=0:5@MAX]
yes? list sst[x=300,y=-20]
VARIABLE : Surface Temperature (deg C)
...
Restricts the scope of the variable name to the named data set. See further discussion in the chapter "Variables and Expressions", section "Defining New Variables".
The qualifier "DATASET=" (LET/DATASET=...) allows you detailed control over the multiple use of the same name.
If the name or number of a data set is supplied then the /dataset qualifier indicates that this variable name is to be defined only in the specified data set. For example
yes? LET/dataset=coads_climatology V_geostrophic = SLP[X=@DDC]/(F*RHO)
Defines V_geostrophic only in data set coads_climatology. In other data sets the name V_geostrophic may refer to file variables or it may be given different definitions or it may be undefined. The data set may be specified either by name as in this example or by number as shown by SHOW DATA. Note that variables defined using LET/dataset=[name_or_number] will be shown in the SHOW DATA output for that data set as well as in SHOW VARIABLES.
If the /dataset qualifier is applied without specifying a data set name then the interpretation is different. In this case the named variable becomes a default definition -- one which applies only if a data-set specific variable of the same name does not exist. For example, if the command
yes? LET/DATASET sst = temp[Z=0]
is issued then sst[D=levitus_climatology] will evaluate to temp[D=levitus_climatology,Z=0] because the variable sst does not exist in levitus_climatology, but sst[D=coads_climatology] will refer to the file variable name sst within the coads_climatology data set.
LET/D is especially useful for editing data sets because it gives a ready way to distinguish between the pre-edit and post-edit versions of the variable. In this example we edit the data set etopo60, replacing a small rectangle in the Pacific Ocean.
Example:
! Do not use memory-cached data when editing. ! Always reread the most recent version from the file. yes? SET MODE STUPID ! Save an exact copy of the original data for editing. ! We will call our edited file "new_etopo.cdf" yes? SET DATA etopo60 yes? LET/D=etopo60 depth = rose yes? SET VARIABLE/TITLE="edited etopo depth"/UNITS=meters depth yes? SAVE/FILE=new_etopo.cdf depth yes? USE new_etopo.cdf ! "rose[d=etopo60]" is the original. ! "depth[d=new_etopo]" is the edited version. ! Redefine "depth[d=etopo60]" as a tool for for selective editing. yes? LET/D=etopo60 depth = rose[D=etopo60]-rose[D=etopo60] + correction ! An example edit: replace a small region with the value 500 yes? LET correction = 500 yes? SAVE/APPEND/FILE=new_etopo.cdf depth[D=etopo60,X=180:175w,Y=0:2n] yes? PLOT/X=160e:160w/Y=1n rose[D=etopo60], depth[D=new_etopo]
This qualifier has no effect. In early versions of Ferret it suppressed a message on redefining a variable. This message is no longer issued.
Specifies a title (in quotation marks) for the user-defined variable. This title will be used to label plots and listings. If no title is specified the text of the expression will be used as the title. (See also SET VARIABLE/TITLE.)
Specifies the units (in quotation marks) of the variable being defined. (See command SET VARIABLE/UNITS.)
/CLIP /ORIGIN /SIZE /TEXT /XLIMITS /YLIMITS/AXES
Defines a new viewport (a sub-rectangle of the graphics window).
yes? DEFINE VIEWPORT[/qualifiers] view_name
Issuing the command SET VIEWPORT is best thought of as entering "viewport mode." While in viewport mode all previously drawn viewports remain on the screen until explicitly cleared with either SET WINDOW/CLEAR or CANCEL VIEWPORT. If multiple plots are drawn in a single viewport without the use of /OVERLAY the current plot will erase and replace the previous one; the graphics in other viewports will be affected only if the viewports overlap. If viewports overlap the most recently drawn graphics will always lie on top, possibly obscuring what is underneath. By default, the state of "viewport mode" is canceled.
Either a comma or a colon separates the upper and lower limits.
Example:
yes? DEFINE VIEWPORT/XLIMITS=0,.5/YLIMITS=0,.5 LL
Defines a viewport that will place graphical output into the lower left quarter of the screen, and names the viewport "LL".
Command qualifiers for DEFINE VIEWPORT.
DEFINE VIEWPORT/XLIMITS=/YLIMITS=
Specifies the portion of the full window to be used.
yes? DEFINE VIEWPORT/XLIMITS=x1,x2/YLIMITS=y1,y2 view_name
The values of the limits must be in the range [0,1]; they refer to the portion of the window (of height and length 1) which defines the viewport. Together, /XLIMITS and /YLIMITS replace the CLIP, ORIGIN, and SIZE qualifiers in older Ferret versions.
Controls shrinkage (or expansion) of text.
yes? DEFINE VIEWPORT/TEXT=n view_name
In some cases text appearance may become unacceptable due to viewport size and aspect specifications. A value of 1 produces text of the same size as in the full window; 0 < n < 1 shrinks the text; n > 1 enlarges text. Sensible values go up to about 2. When the qualifier /TEXT is omitted, Ferret computes a text size that is appropriate to the size of the viewport.
Note that /TEXT modifies the prominence of the text through manipulation of axis lengths rather than through direct manipulation of the many text size specifications. A low value of text prominence produces axes that are "long" (as seen with SHOW SYMBOLS, or PPL LIST XAXIS), making the (fixed size) text appear less prominent.
DEFINE VIEWPORTS/AXES Specifies that user's limits are specifying where the plot axes lie as a fraction of the entire plot window. Either a comma or a colon separates the upper and lower limits.
DEFINE VIEWPORT/XLIMITS=x1,x2/YLIMITS=y1,y2/AXES view_name
Example 1:
These commands define two viewports, entirely filling the upper two quadrants of the plot window:
yes? DEFINE VIEWPORT/AXES/XLIM=0:0.5/YLIM=0.5:1 ulax yes? DEFINE VIEWPORT/AXES/XLIM=0.5:1/YLIM0=.5:1 urrax yes? set view ulax yes? shade/nokey/i=1:5/y=1:5 i+j yes? set view urax yes? shade/nokey/i=1:5/y=1:5 i+j
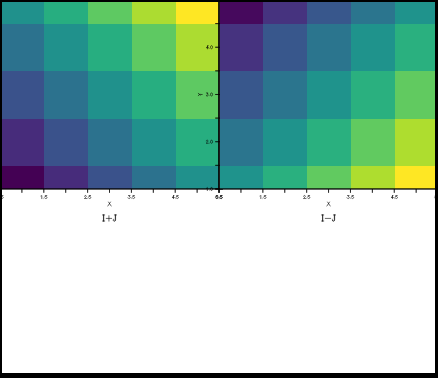
Plot elements that lie outside the axes are still drawn, for instance colorbars, or the labels along plot axes, as seen above.
When a viewport is defined along the lower edge of the window PPLUS will add space automatically to allow for labels which would otherwise be lost. This behavior can be changed using PPL commands to set the plot origin, or the qualifier /NOYADJUST can be added to the plot command to draw the axes at the locations specified in the viewport definition.
Example 2:
This example shows the effect of the /NOYADJUST qualifier on the plot command. Define two viewports and plot; on the left the Y axis is adjusted automatically, on the right we specify /NOADJUST and the labelling below the plot is not plotted.
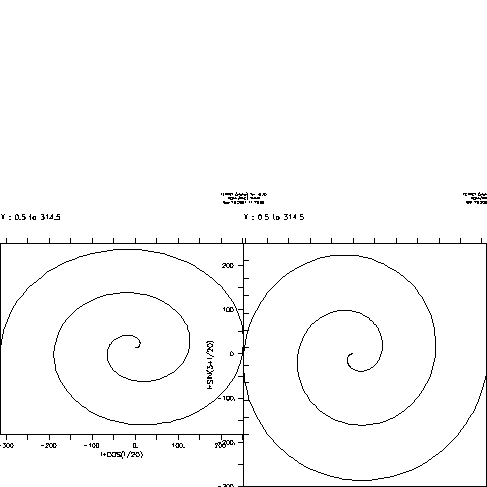
yes? DEFINE VIEW/AXES/XLIM=0:0.5/YLIM=0:0.5 llax yes? DEFINE VIEW/AXES/XLIM=0.5:1/YLIM=0:0.5 lrax yes? SET VIEW llax yes? PLOT/VS/LINE/I=1:314 i*cos(i/20),i*sin(i/20) yes? SET VIEW lrax yes? PLOT/VS/LINE/I=1:314/NOYADJUST i*cos(3+i/20),i*sin(3+i/20)
Example 3:
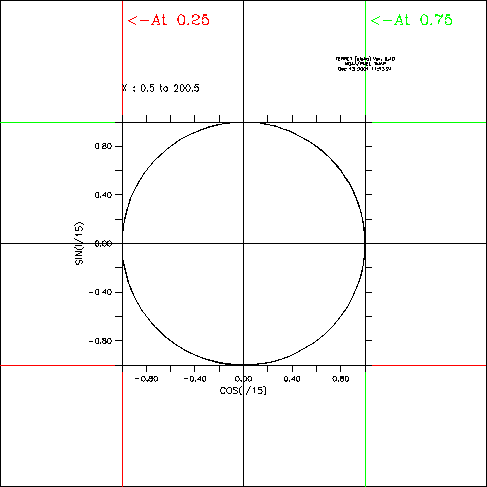
DEFINE VIEWPORT/AXES can be used to set guide lines on a plot
yes? CANCEL VIEW
yes? DEFINE VIEW/AXES allax
yes? SET VIEW allax
yes? PLOT/VS/LINE/HLIM=0:1/VLIM=0:1/NOLAB {0.5,0.5,,0,1},{0,1,,0.5,0.5}
yes? PLOT/VS/LINE/OVER/NOLAB {0.25,0.25,,0,1},{0,1,,0.25,0.25}
yes? PLOT/VS/LINE/OVER/NOLAB {0.75,0.75,,0,1},{0,1,,0.75,0.75}
yes? LABEL 0.26,0.95,-1,0,.2 @P2@AC<-At 0.25
yes? LABEL 0.76,0.95,-1,0,.2 @P3@AC<-At 0.75
yes? DEFINE VIEW /XLIM=0.25:0.75/YLIM=0.25:0.75/TEXT=1/AXES mid
yes? SET VIEW mid
yes? PLOT/VS/HLIM=-1:1/VLIM=-1:1/LINE/I=1:200 cos(i/15),sin(i/15)
DEFINE VIEWPORT/CLIP=
This qualifier is obsolete; see XLIMITS= and /YLIMITS= (above). Specifies the location of the upper right corner of the viewport.
DEFINE VIEWPORT/ORIGIN=
This qualifier is obsolete; see /XLIMITS= and /YLIMITS= (above). Specifies the location of the lower left corner of the viewport.
DEFINE VIEWPORT/SIZE=
This qualifier is obsolete; see /XLIMITS and /YLIMITS (above). Specifies the scaling factor to use relative to the size of the full window.
DEFINE DATA/AGGREGATE /T /E /F /U /TITLE /QUIET /HIDE /TAXIS /FAXIS /TOVERLAP name = [list of datsets]
Defines a virtual dataset, aggregating a set of datsets. This command is implemented only in the T, E, and F directions. In addition DEFINE DATA/U defines a Union aggregation.
TSERIES is an alias for DEFINE DATA/AGGREGATE/T
Define an time series dataset from a set of datasets that represent timesteps or sets of timesteps, containing those variables from the datasets that have identical variable names and grids. This command is in many ways the equivalent of using a descriptor file or multi-CDF dataset. Each method for creating a virtual dataset has advantages. Multi-file datasets will initialize more quickly than making an aggregation definition, as the "DEFINE DATA" step needs to initialize all of the member datasets to determine the time axis, but in a multi-CDF dataset, that information is read from the descriptor file. With a TSERIES aggregation, however, the correct axis bounds are applied, and missing time steps are handled by filling the data for those time steps with missing-value flags. Also a TSERIES aggregation is more flexible, making a time series of the time-step datasets which are currently available.
ENSEMBLE is an alias for DEFINE DATA/AGGREGATE/E
Define an ensemble dataset from a set of datasets, containing those variables from the datasets that have identical variable names and grids.
FMRC is an alias for DEFINE DATA/AGGREGATE/F
Define a forecast/time dataset (FMRC, for Forecast Model Run Collection), from a set of individual forecast datasets run for a set of times; containing those variables from the datasets that have identical variable names and grids.
UNION is an alias for DEFINE DATA/AGGREGATE/U
Define a Union aggregation, from a set of files that contain different variables on a common grid.
DEFINE DATA is valid only with /AGGREGATE. It specifies that we are defining a virtual dataset by aggregation.
The syntax "use my_definition.agg" will run a Ferret script containing an aggregation definition, as a convenience. The users is responsible for the contents of the script; all that is happening is that Ferret translates "use " to "go " when the file extension is ".agg".
The extension .agg may be left off; it is added to the file extensions for datasets that are handled automatically, and the dataset directories will be searched for the .agg file.
Define the aggregate in the T direction. The variables in the member datsets must have a T axis with the same time units. The time origins do not need to match. Ferret computes correct common time steps for the aggregated time axis. The DEFINE DATA command will define a time axis, and the variables which are common to all of the member datasets will be listed on this time axis. Any variables without a time axis in member datasets will be included in the aggregated dataset. The list of datasets need not be ordered in time; Ferret will reorder the members in time.
If the list of files comes from a string variable, or from a spawn command, the list does not need to be ordered by time. Ferret will order the members of the files in time.
NOTE: For time aggregations, we are aggregating netCDF files or URL's, not Ferret datasets. The list of datasets must be the file names with path, or the url's, not Ferret dataset numbers.
Define the aggregate in the E direction. The variables in the member datsets must not have an E axis. The DEFINE DATA command will define an abstract E axis, and the members of the aggregation will be listed on this axis in the order they are given on the command line.
For ensembles defined on the fly, a variable is defined along the E axis which contain the dataset names. Variables containing global attributes are also promoted, so for instance an attribute with the model_name becomes a variable in the virtual dataset. These variables promoted from the ensemble members are created in versions 7.42 and higher. See examples below.
If you save data on an ensemble axis to a file, the Time axis in the saved file is saved as a fixed time axis, not (as is usually the default) an unlimited-coordinates axis that could be appended to. In order to append in time, the file will need to be created on the first write to it, with a /LLIMITS command defining the full time length to be written using any subsequent /SAVE/APPEND commands. (See LIST/LLIMITS /MLIMITS and LIST/RIGID for more information.)
Define the aggregate in the F direction. The variables in the member datsets must not have an F axis. The DEFINE DATA/AGG/F command will define a 2-dimensional Time/Forecast coordinate variable, which it names TF_TIMES. For more about working with these datasets, see the secion about FMRC datasets in Chapter 4.
If you save data on an F axis to a file, the Time axis in the saved file is saved as a fixed time axis, not (as is usually the default). In order to append in time, the file will need to be created on the first write to it, with a /LLIMITS command defining the full time length to be written using any subsequent /SAVE/APPEND commands. (See LIST/LLIMITS /MLIMITS and LIST/RIGID for more information.)
Define a Union aggregation. Variables in the member datsets must be on the same grid.
Set an optional title for the new dataset, which appears on plots and listings as the dataset title.
Suppresses warning messages about variables in the member datasets which do not share grids or are not present in all member datsets.
Marks the member datasets as "hidden". This means that SHOW DATA commands will not list the member datasets but only the ensemble datasets (and any other datasets that may be open but are not in an ensemble). SHOW DATA/HIDDEN will show all datasets, even ones that are hidden; and SHOW DATA with a dataset number or name will show that dataset, whether or not it is hidden.
For Time aggregations only, the desired time axis may be pre-defined and passed to the DEFINE DATA command. The time coordinates in the files are checked against the times in the axis. The times in the member datasets are sorted, just as they are without /TAXIS=.
Those times that agree exactly are matched with those on the time axis; if there are gaps in the times in the member datasets those gaps are represented by missing-data flags in the aggregation.
yes? ! Aggregate these datasets - may have gaps before, after, and
yes? ! between when viewed along mytax
yes? LET aggfiles = { "member_02.nc", "member_13.nc", "member_24.nc", "member_35.nc" }
yes? DEFINE DATA /AGGREGATE /HIDE /T /TAXIS=mytax timeagg = aggfiles
*** NOTE: Time axis coordinates matched using @XACTIf the timesteps in the member files do not exactly match those on the specified time axis, but the times represented can be found along the axis, they are mapped to the time axis using a binning operation.
yes? DEFINE DATA /AGGREGATE /HIDE /T /TAXIS=tax timeagg = aggfiles
*** NOTE: Time axis coordinates matched using @BINIf the timesteps in the member files do not correspond to the defined axis, the files are simply assigned to the axis using an @ASN transformation. If there are more timesteps in the axis than in the member datasets, missing-data fills in the end of the timeseries.
yes? DEFINE DATA /AGGREGATE /HIDE /T /TAXIS=tax2 timeagg = aggfiles
*** NOTE: Time axis coordinates matched using @ASNDEFINE DATA/T/TAXIS= /TOVERLAP[=max]
For time aggregations only, /TOVERLAP may be specified in combination with /TAXIS= to allow timesteps in the member datasets to overlap. The timesteps at the start of each member set be used in the aggregation if they overlap timesteps from a previous member. If an argument is given, then the aggregation is defined only if the number of timesteps of overlap is "max".
yes? ! Files are ordered in time and overlaps found and noted.
yes? DEFINE DATA /AGG /T /TOVERLAP /TAXIS=aggtime myagg = d2010.nc, d2009.nc, d2011.nc
*** NOTE: Ignored last 1 timesteps of d2009.nc due to overlap with d2010.nc
*** NOTE: Ignored last 3 timesteps of d2010.nc due to overlap with d2011.nc
*** NOTE: Time axis coordinates matched using @XACT
yes? ! Only do the aggregation if the overlap is 2 or fewer timesteps.
yes? DEFINE DATA /AGG /T /TOVERLAP=2 /TAXIS=aggtime@XACT agg2 = d2010.nc, d2009.nc, d2011.nc
*** NOTE: Ignored last 1 timesteps of d2009.nc due to overlap with d2010.nc
**ERROR: error defining aggregate dataset: Excessive (3) overlaps in time axes for d2010.nc and d2011.nc
The dataset name may be any valid name under Ferret naming syntax.
The list of datasets may be a set of dataset numbers referring to datasets that have already been initialized. Or they may be any valid NetCDF file/path names or OPeNDAP dataset names. The datasets may be listed in a string valued array, or may be specified using the results of a SPAWN command. If the datasets are not currently open, the DEFINE DATA command will open them.
For a TSERIES aggregation, the list of datasets must be the NetCDF file/path names or OPeNDAP dataset names and may NOT be the dataset numbers of datasets that may be open in Ferret. For time aggregations the list of datasets does not need to be ordered in time; the correct time ordering will be found as the aggregation is created.
Examples:
yes? DEFINE DATA/AGGR/E/TITLE="Pacific Ocean model ensemble" PACMOD = 1, 2, 3, 4 yes? ENSEMBLE/TITLE="Pacific Ocean model ensemble" PACMOD = 1, 2, 3, 4 ! equivalent to the above yes? DEFINE DATA/AGGR/F/TITLE="Climate FMRC" fmrdat = 1,2,3,4,5,6,7,8,9,10 yes? FMRC TITLE="Climate FMRC" fmrdat = 1,2,3,4,5,6,7,8,9,10 ! equivalent to the above
! Time series aggregations may be specified only using file or URL names
! or a variable that lists the names
yes? define data/agg/t myagg2 = /home/data/tagg_reg_1.nc, /home/data/tagg_reg_2.nc
! or
yes? let file_list = SPAWN("ls -1 tmp/tagg_set_?.nc")
yes? TSERIES listAgg = file_list
yes? show data/members listAgg
yes? USE ens1.nc yes? USE ens2.nc yes? USE ens3.nc yes? DEFINE DATA/AGGR/E/TITLE="3-dataset ensemble" ens3 = 1, 2, 3 yes? ENSEMBLE ocean_agg = "http://server1.address/directory/model_A.nc",\ "http://another_server/directory/subdirectory/model_B.nc"
yes? use run1.nc
yes? use run2.nc
yes? use run3.nc
yes? use run4.nc
yes? FMRC/hide all_runs = 1,2,3,4
yes? FMRC/hide nmme_2 = SPAWN("cat ($urlfile)")
yes? show data/members
yes? show data 5.2 ! lists details of member 2 of datset 5 (the FMRC dataset)
yes? UNION uset = tmp/sst.nc,tmp/uwnd.nc,tmp/vwnd.nc,tmp/airt.nc
SHOW DATA/MEMBERS agg_set_name will show the aggregation and its members, listing the timesteps of the members for an aggregation in time
Ferret checks that the datasets are open and looks at all of the variables. If the datasets share a variable, it checks that their grids are identical. Units of variables and other attributes are NOT checked. If a variable is not in all datasets, or if grids do not match, it is not included in the dataset. The units of the variable in the first dataset will be used in the aggregated set. Use SET VAR/UNITS, SET VAR/TITLE, etc to reset any units etc. that you wish to change.
The Ferret default missing-data flag is used for the aggregated variables. The missing-data flag for each variable in its source dataset is applied when that variable is read as a member of an ensemble dataset, so these flags do not need to match for variables coming from different datasets.
Aggregations may be aggregated. For instance, a set of TSERIES aggregations may then be aggregated in Ensemble direction; and those combined into a Forecast. Or aggregations in T, E, and/or F may then be combined as a UNION aggregation.
ENSEMBLE Examples:
Say that these datasets all share a variable SST, but only the first also contains AIRT. Note how the ensemble set has just SST of the file variables, and it has variables promoted from the global attributes of the member datasets: The member dataset names, the history attribute, the title attribute, the model title, etc.
Example 1:
yes? USE ens1, ens2, ens3, ens4
! list the global attributes of the first dataset
yes? set data 1
yes? show attributes .
attributes for dataset: ./data/ens1.nc
..title = Example datset to be used in aggregations
..history = FERRET V7.4 (optimized) 16-May-18
..Conventions = CF-1.6
..MODEL = modelA
..NUMBER = 1
yes? ENSEMBLE/title="Ensemble of three" threefiles = ens1, ens2, ens3
*** NOTE: Exclude variable from aggregate. Does not appear in all member datasets: AIRT
yes? sh dat
currently SET data sets:
1> ./ens1.nc
name title I J K L M N
SST SST_IN 1:10 1:9 ... 1:12 ... ...
AIRT AIR TEMPERATURE 1:10 1:9 ... 1:12 ... ...
2> ./ens2.nc
name title I J K L M N
SST SST_IN 1:10 1:9 ... 1:12 ... ...
3> ./ens3.nc
name title I J K L M N
SST SST_IN 1:10 1:9 ... 1:12 ... ...
4> ./ens4.nc
name title I J K L M N
SST SST_IN 1:10 1:9 ... 1:12 ... ...
5> threefiles (default) Ensemble aggregation
name title I J K L M N
SST SST_IN 1:10 1:9 ... 1:12 1:3 ...
------------------------------
MEMBER_DSET
Member dataset names in Ensembl ... ... ... ... 1:3 ...
TITLE Global Att *title* in Ensemble ... ... ... ... 1:3 ...
HISTORY Global Att *history* in Ensembl ... ... ... ... 1:3 ...
CONVENTIONS
Global Att *Conventions* in Ens ... ... ... ... 1:3 ...
MODEL Global Att *MODEL* in Ensemble ... ... ... ... 1:3 ...
NUMBER Global Att *NUMBER* in Ensemble ... ... ... ... 1:3 ...
yes? list model
VARIABLE : Global Att *MODEL* in Ensemble
DATA SET : Ensemble of three
FILENAME : threefiles
SUBSET : 3 points (E (realization))
1 / 1:"modelA"
2 / 2:"modelB"
3 / 3:"modelC"
yes? cancel data/all
yes? define data/agg/E/hide fourfiles = ens1, ens3, ens2, ens4
*** NOTE: Variable "AIRT" excluded from aggregate: Not found in all member datasets
yes? show data/members fourfiles
currently SET data sets:
5> fourfiles (default) Ensemble aggregation
name title I J K L M N
SST SST_IN 1:10 1:9 ... 1:12 1:4 ...
------------------------------
MEMBER_DSET
Member dataset names in Ensembl ... ... ... ... 1:4 ...
TITLE Global Att *title* in Ensemble ... ... ... ... 1:4 ...
HISTORY Global Att *history* in Ensembl ... ... ... ... 1:4 ...
CONVENTIONS
Global Att *Conventions* in Ens ... ... ... ... 1:4 ...
MODEL Global Att *MODEL* in Ensemble ... ... ... ... 1:4 ...
NUMBER Global Att *NUMBER* in Ensemble ... ... ... ... 1:4 ...
Member datasets:
5.1: ./data/ens1.nc
5.2: ./data/ens3.nc
5.3: ./data/ens2.nc
5.4: ./data/ens4.nc
yes? cancel data fourfiles
! Canceling the ensemble closes all of the datasets
yes? show data/hidden
currently SET data sets:
If the datasets do not share any variables on matching grids, the ensemble dataset is not created. If one or more datasets cannot be opened, the dataset is not created.
Example 2:
yes? use coads_climatology yes? use levitus_climatology yes? define data/agg/e/title="two climatologies" clim_e = 1,2 *** NOTE: Exclude variable from aggregate. Does not appear in all member datasets: SST **ERROR: error defining aggregate dataset: No valid datasets or datasets share no variables.
To define an ensemble where the variable names or grids do not match, define variables in one or more of the datasets, putting them onto the same grid and ensuring that they have the same name. Use the "DEFINE VARIABLE/DATASET" syntax to define a new variable associated with a particular dataset. Then this variable will be included in the search for matching variables in the member datasets. NOTE: At least one file must contain each of the variables being aggregated in the ensemble, as a file variable not a LET/D variable. See example 4 below.
Example 3: Create an ensemble dataset with UWND and VWND from coads_climatology and monthly_navy_winds datasets. In the original files these variables have different grids in X, Y, and T.
yes? use coads_climatology
yes? use monthly_navy_winds
yes? sh dat
currently SET data sets:
1> /home/users/tmap/ferret/linux/fer_dsets/data/coads_climatology.cdf
name title I J K L M N
SST SEA SURFACE TEMPERATURE 1:180 1:90 ... 1:12 ... ...
AIRT AIR TEMPERATURE 1:180 1:90 ... 1:12 ... ...
SPEH SPECIFIC HUMIDITY 1:180 1:90 ... 1:12 ... ...
WSPD WIND SPEED 1:180 1:90 ... 1:12 ... ...
UWND ZONAL WIND 1:180 1:90 ... 1:12 ... ...
VWND MERIDIONAL WIND 1:180 1:90 ... 1:12 ... ...
SLP SEA LEVEL PRESSURE 1:180 1:90 ... 1:12 ... ...
2> /home/users/tmap/ferret/linux/fer_dsets/data/monthly_navy_winds.cdf (default)
name title I J K L M N
UWND ZONAL WIND 1:144 1:73 ... 1:132 ... ...
VWND MERIDIONAL WIND 1:144 1:73 ... 1:132 ... ...
Let's put the monthly winds data onto the same climatological time axis and XY grid as coads_climatology. We want to keep the names of the original variables UWND and VWND, so rename the file variables to uwnd_in, vwnd_in in the second dataset.
! Rename the file variables, so we can use UWND and VWND for our new variable names. yes? SET VAR/NAME=uwnd_in uwnd[d=2] yes? SET VAR/NAME=vwnd_in vwnd[d=2]
Now define UWND and VWND, regridding them onto axes from coads_climatology. The time axis in dataset 1 is a modulo time axis, so use @MOD regridding.
yes? LET/UNITS="M/S"/TITLE="Zonal Wind"/D=2 UWND = uwnd_in[D=2,GXY=uwnd[d=1],GT=uwnd[d=1]@MOD] yes? LET/UNITS="M/S"/TITLE="Meridional Wind"/D=2 VWND = vwnd_in[D=2,GXY=vwnd[d=1],GT=vwnd[d=1]@MOD]
See the variables we defined.
yes? SHOW DATA
currently SET data sets:
1> /home/users/tmap/ferret/linux/fer_dsets/data/coads_climatology.cdf
name title I J K L
SST SEA SURFACE TEMPERATURE 1:180 1:90 ... 1:12
AIRT AIR TEMPERATURE 1:180 1:90 ... 1:12
SPEH SPECIFIC HUMIDITY 1:180 1:90 ... 1:12
WSPD WIND SPEED 1:180 1:90 ... 1:12
UWND ZONAL WIND 1:180 1:90 ... 1:12
VWND MERIDIONAL WIND 1:180 1:90 ... 1:12
SLP SEA LEVEL PRESSURE 1:180 1:90 ... 1:12
2> /home/users/tmap/ferret/linux/fer_dsets/data/monthly_navy_winds.cdf (default)
name title I J K L M N
UWND_IN ZONAL WIND 1:144 1:73 ... 1:132 ... ...
VWND_IN MERIDIONAL WIND 1:144 1:73 ... 1:132 ... ...
------------------------------
VWND[D=monthly_navy_winds] = VWND_IN[D=2,GXY=VWND_IN[D=1],GT=VWND_IN[D=1]@MO]
UWND[D=monthly_navy_winds] = UWND_IN[D=2,GXY=UWND_IN[D=1],GT=UWND_IN[D=1]@MOD]
The uwnd and vwnd in both datatsets have the same axes.
yes? sh grid uwnd[d=1]
GRID GSQ1
name axis # pts start end
COADSX LONGITUDE 180mr 21E 19E(379)
COADSY LATITUDE 90 r 89S 89N
normal Z
TIME TIME 12mr 16-JAN 06:00 16-DEC 01:20
normal E
normal F
yes? sh grid uwnd[d=2]
GRID GSQ1
name axis # pts start end
COADSX LONGITUDE 180mr 21E 19E(379)
COADSY LATITUDE 90 r 89S 89N
normal Z
TIME TIME 12mr 16-JAN 06:00 16-DEC 01:20
normal E
normal F
Now, we have variables UWND and VWND in both datasets and they have the same grids. We can define an aggregation. (Here's where /QUIET could be used, to supress all these NOTE's.) Because we use /HIDE on the define data command, SHOW DATA lists only the ensemble dataset.
! Use ENSEMBLE/QUIET to suppress all of the *** NOTE lines
yes? ENSEMBLE/HIDE/TITLE="two winds" windy = 1,2
*** NOTE: Variable "SST" excluded from aggregate: Not found in all member datasets
*** NOTE: Variable "AIRT" excluded from aggregate: Not found in all member datasets
*** NOTE: Variable "SPEH" excluded from aggregate: Not found in all member datasets
*** NOTE: Variable "WSPD" excluded from aggregate: Not found in all member datasets
*** NOTE: Variable "SLP" excluded from aggregate: Not found in all member datasets
yes? SHOW DATA
currently SET data sets:
3> WINDY (default) Ferret-defined Ensemble dataset
name title I J K L M N
UWND ZONAL WIND 1:180 1:90 ... 1:12 1:2 ...
VWND MERIDIONAL WIND 1:180 1:90 ... 1:12 1:2 ...
------------------------------
MEMBER_DSET
Member dataset names in Ensembl ... ... ... ... 1:2 ...
HISTORY Global Att *history* in Ensembl ... ... ... ... 1:2 ...
yes? ! To see the member datasets, use SHOW DATA/HIDDEN
Now we have the variables and can work with them in the ensemble dimension.
yes? vec/L=3 uwnd[M=@ave], vwnd[M=@ave]
If that seems like a lot of work for something we could have done with other Ferret commands, imagine if there were 10 datasets that we want to treat as an ensemble. Most often these will be a set of model runs with variables and grids that already match. Having the E axis along which we can do operations is the key here.
Example 4: One file has temperature data on an x-y-z-t grid; the others have surface temperature only. Define a LET/D variable in file 1, and define the aggregation. So long as each variable exists as a FILE variable (as opposed to a user-defined variable) in at least one member set, we can define the ensemble to include that variable.
yes? use a, b, c
yes? show data
currently SET data sets:
1> ./a.nc
name title I J K L
TEMP Temperature 1:180 1:90 1:34 1:12
2> ./b.nc
name title I J K L
SST SEA SURFACE TEMPERATURE 1:180 1:90 ... 1:12
3> ./c.nc (default)
name title I J K L
SST SEA SURFACE TEMPERATURE 1:180 1:90 ... 1:12
yes? let/d=1/TITLE="SEA SURFACE TEMPERATURE" sst = temp[K=1]
yes? ENSEMBLE surface_temp = a, b, c
*** NOTE: Variable "TEMP" excluded from aggregate: Not found in all member datasets
yes? SHOW DATA surface_temp
currently SET data sets:
4> surface_temp (default) Ensemble aggregation
name title I J K L M N
SST SEA SURFACE TEMPERATURE 1:180 1:90 ... 1:12 1:3 ...
------------------------------
MEMBER_DSET
Member dataset names in Ensembl ... ... ... ... 1:3 ...
HISTORY Global Att *history* in Ensembl ... ... ... ... 1:3 ...
CONVENTIONS
Global Att *Conventions* in Ens ... ... ... ... 1:3 ...
For further examples using Ensemble datasets, see the PDFs
PyFerret_exploring_ensembles.pdf
PyFerret_ensembles_of_FMRCs.pdf
TSERIES examples:
yes? define data/agg/t myagg8 = tmp/reg_1.nc, tmp/reg_2.nc,tmp/reg_3.nc, tmp/reg_4.nc,tmp/reg_5.nc
yes? show data/members
currently SET data sets:
1> myagg5 (default)
name title I J K L M N
MYVAR SIN(T[GT=TDAY]/10) + L[GT=TDAY] ... ... ... 1:50 ... ...
MYMAP I[X=0E:0E:5]+ 10*J[Y=80S:80N:5] 1:73 1:33 ... ... ... ...
time-dependent data files:
01-JAN-1980 00:00 -> 10-JAN-1980 00:00 tmp/tagg_reg_1.nc
11-JAN-1980 00:00 -> 20-JAN-1980 00:00 tmp/tagg_reg_2.nc
21-JAN-1980 00:00 -> 30-JAN-1980 00:00 tmp/tagg_reg_3.nc
31-JAN-1980 00:00 -> 09-FEB-1980 00:00 tmp/tagg_reg_4.nc
10-FEB-1980 00:00 -> 19-FEB-1980 00:00 tmp/tagg_reg_5.nc
A common situation is datasets which have one timestep per file. Here, we have daily data, but day 3 is missing:
yes? TSERIES single_irreg = tmp/day_1.nc, tmp/day_2.nc, tmp/day_4.nc, tmp/day_5.nc
yes? show data single_irreg
currently SET data sets:
1> single_irreg (default)
name title I J K L M N
MYVAR SIN(T[GT=TDAY]/10) + L[GT=TDAY] ... ... ... 1:4 ... ...
yes? list myvar
VARIABLE : SIN(T[GT=TDAY]/10) + L[GT=TDAY]/10
FILENAME : single_irreg
SUBSET : 4 points (TIME)
01-JAN-1980 00 / 1: 0.1179
02-JAN-1980 00 / 2: 0.3177
04-JAN-1980 00 / 3: 0.7126
05-JAN-1980 00 / 4: 0.9059
When using a list of files to define the aggregation, the directory listing may not be correctly ordered in time. The time aggregation takes care of finding the correct order.
yes? let file_list = SPAWN("ls -1 tmp/step_*.nc")
yes? list file_list
VARIABLE : SPAWN("ls -1 tmp/step_*.nc")
SUBSET : 4 points (X)
1 / 1:"tmp/step_12.nc"
2 / 2:"tmp/step_18.nc"
3 / 3:"tmp/step_24.nc"
4 / 4:"tmp/step_6.nc"
yes? define data/agg/t steps = file_list
yes? sh dat/members steps
currently SET data sets:
1> steps (default)
name title I J K L M N
MYVAR SIN(T[GT=TDAY]/10) + L[GT=TDAY] ... ... ... 1:4 ... ...
time-dependent data files:
06-JAN-1980 00:00 -> 06-JAN-1980 00:00 tmp/step_6.nc
12-JAN-1980 00:00 -> 12-JAN-1980 00:00 tmp/step_12.nc
18-JAN-1980 00:00 -> 18-JAN-1980 00:00 tmp/step_18.nc
24-JAN-1980 00:00 -> 24-JAN-1980 00:00 tmp/step_24.nc
yes? list myvar
VARIABLE : SIN(T[GT=TDAY]/10) + L[GT=TDAY]/10
FILENAME : steps
SUBSET : 4 points (TIME)
06-JAN-1980 00 / 1: 1.095
12-JAN-1980 00 / 2: 2.099
18-JAN-1980 00 / 3: 2.789
24-JAN-1980 00 / 4: 3.134
FMRC examples. The aggregation defines the 2-dimensional Forecast time step matrix for the dataset. It is automatically named TF_TIMES.
! define a complete FMRC aggregation
yes? let files = SPAWN("cat file_list.dat")
yes? list files ! notice that they are not properly ordered
VARIABLE : SPAWN("ls -1 tmp/fcst_*.nc")
SUBSET : 11 points (X)
1 / 1:"tmp/fcst_10.nc"
2 / 2:"tmp/fcst_11.nc"
3 / 3:"tmp/fcst_1.nc"
4 / 4:"tmp/fcst_2.nc"
5 / 5:"tmp/fcst_3.nc"
6 / 6:"tmp/fcst_4.nc"
7 / 7:"tmp/fcst_5.nc"
8 / 8:"tmp/fcst_6.nc"
9 / 9:"tmp/fcst_7.nc"
10 / 10:"tmp/fcst_8.nc"
11 / 11:"tmp/fcst_9.nc"
yes? fmrc my_fmrc = files
yes? show data/brief ! note that member files are hidden
currently SET data sets:
12> my_fmrc (default) Forecast aggregation
yes? show data/members my_fmrc ! note that the ordering is correct.
currently SET data sets:
12> my_fmrc (default) Forecast aggregation
name title I J K L M N
FCST FIELD + FCST_ERROR 1:21 1:21 ... 1:24 ... 1:11
(L=1:24)
TF_TIMES Forecast time step matrix ... ... ... 1:24 ... 1:11
FCST_2 another variable 1:21 1:21 ... 1:24 ... 1:11
(L=1:24)
Aggregated datasets:
1 : tmp/fcst_1.nc
2 : tmp/fcst_2.nc
3 : tmp/fcst_3.nc
4 : tmp/fcst_4.nc
5 : tmp/fcst_5.nc
6 : tmp/fcst_6.nc
7 : tmp/fcst_7.nc
8 : tmp/fcst_8.nc
9 : tmp/fcst_9.nc
10: tmp/fcst_10.nc
11: tmp/fcst_11.nc
! Show the forecast variable on the native, compact T-F grid
yes? show grid fcst
GRID (G015) Forecast Aggregation Grid
name axis # pts start end subset
XAX LONGITUDE 21mr 140E 160E full
YAX LATITUDE 21 r 20N 40N full
normal Z
TF_LAG_T MODEL ELAPSED TIME (24 r 15.196 714.2 full
normal E
TF_CAL_F FORECAST 11 i 01-JAN-1950 00:00 01-JUL-1952 00:00 full
yes? shade/i=10/j=10 fcst
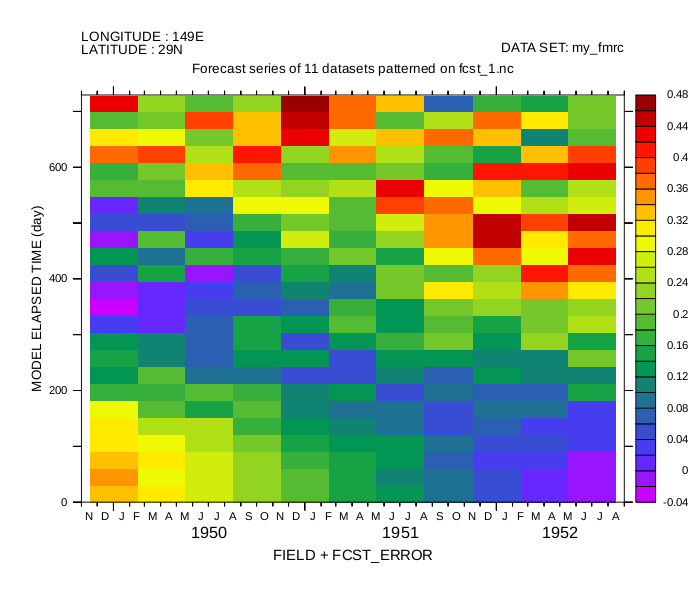
Diagonal form, using Auxiliary-variable regridding to apply the information in the TF_TIMES coordinate matrix.
yes? shade/i=10/j=10 fcst[gt(tf_times)=TF_CAL_T@FMRC]
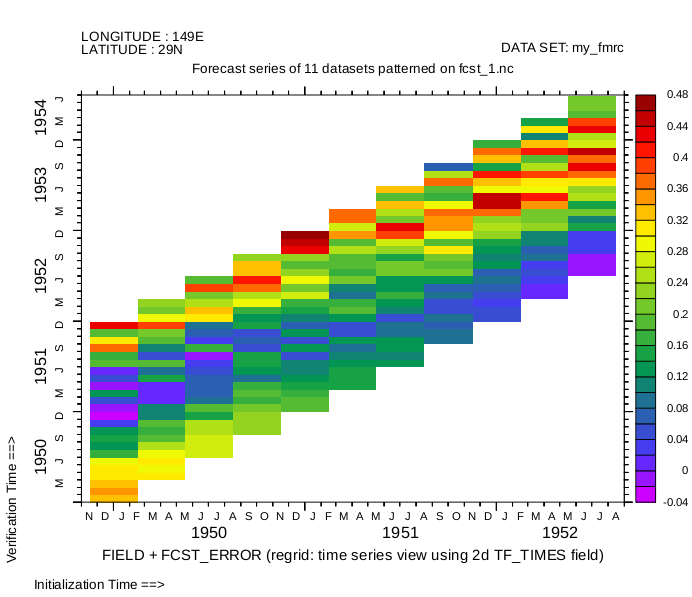
For further examples using FMRC datasets, see these documents:
Using_Forecast_Model_Run_Collections_in_Ferret.pdf
PyFerret_ensembles_of_FMRCs.pdf
Pyferret only
DEFINE PYFUNC [ /NAME=alias ] module.name
Creates an external function using the Python module given by module.name. If /NAME is given, the name of the function will the the name given by alias.